 A photo of me in Oboke, Shikoku, Japan.
A photo of me in Oboke, Shikoku, Japan.Siyuan Stella Wang's Homepage
I am a postdoctoral researcher working in the lab of Dr. William Shih at the Wyss Institute for Biologically Inspired Engineering at Harvard University. I am currently working on making crisscross origami structures bigger and better with surface-supported assembly and developing high-throughput single-molecule protein characterization devices using DNA nanotechnology.
I obtained my PhD in Cell and Molecular Biology from the University of Texas at Austin in 2022, where I was mentored by Dr. Andrew Ellington. Prior to my doctoral studies, I completed my undergraduate studies at the California Institute of Technology, where I earned my degree in Biomolecular Engineering under the guidance of my advisor Dr. Lulu Qian.
Curriculum Vitae ORCID LinkedIn ThesisResearch at the Wyss Institute
 The solid-phase assembly workflow for assembling crisscross origami
The solid-phase assembly workflow for assembling crisscross origamiSSW, Matthew Aquilina, Florian Katzmeier, Huangchen Cui, Jaewon Lee, William Shih. Solid-phase assembly of micrometer-scale crisscross DNA origami structures.
Poster at DNA31 (31st International Conference on DNA Computing and Molecular Programming, August 25-28, 2025, Lyon, France)Matthew Aquilina, Florian Katzmeier, SSW, Corey Becker, Huangchen Cui, Minke Nijenhuis, Yichen Zhao, Su Hyun Seok, William Shih. #-CAD: Software suite for the automated design of crisscross DNA origami.
GitHubSamuel Leitao, Andrew Ward, SSW, William Shih, Wesley Wong. Single-molecule mechano-proteomics with DNA nanoswitch caliper trains.
Collaboration with researchers at the University of Fribourg. Assembly of semiconducting carbon nanotube arrays with DNA crisscross origami.
Collaboration with researchers at the Columbia University. Crisscross origami-templated neuromorphic computing devices self-assembled from 3D optical metamaterials.
Collaboration with researchers at Aarhus University and the Technical University of Denmark. Self-assembly of a bio-nano-wire device on a microelectric chip for single-molecule biosensing and conductance analysis.
Collaboration with researchers at Korea University. Development of a DNA Quantum Foundry for engineering DNA quantum reservoir computers.
Graduate Research
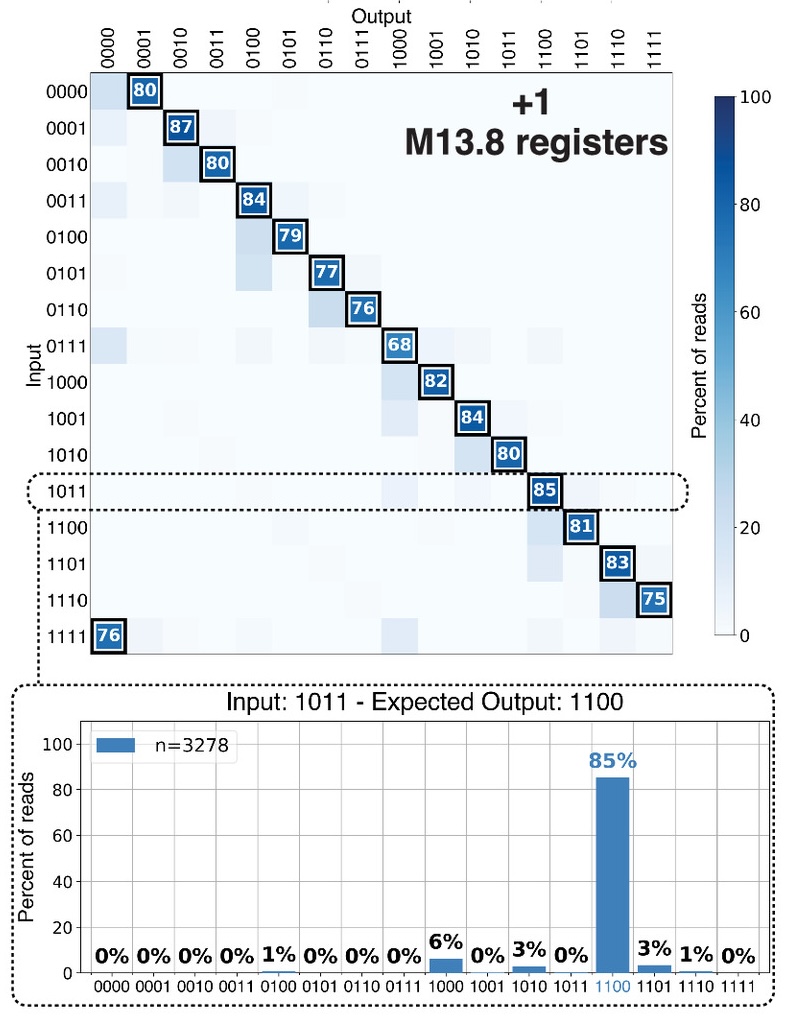 Resulting data of a binary counting algorithm implemented with DNA strand displacement and recorded on ssDNA M13 bacteriophage plasmids
Resulting data of a binary counting algorithm implemented with DNA strand displacement and recorded on ssDNA M13 bacteriophage plasmidsBoya Wang, SSW, Cameron Chalk, Andrew D. Ellington, David Soloveichik. Parallel molecular computation on digital data stored in DNA. Published September 5, 2023
GitHub doi:10.1073/pnas.2217330120SSW, Erhu Xiong, Sanchita Bhadra, Andrew D. Ellington. Developing predictive hybridization models for phosphorothiolated oligonucleotides using high-resolution melting. Published May 18, 2022
doi:10.1371/journal.pone.0268575Raghav Shroff, Jared Ellefson, SSW, Alexander Boulgakov, Randall A. Hughes, Andrew D. Ellington. Recovery of information stored in modified DNA with an evolved polymerase. Published February 3, 2022
doi:10.1021/acssynbio.1c00575SSW, Andrew D. Ellington. Pattern Generation with Nucleic Acid Chemical Reaction Networks. Published March 13, 2019
doi:10.1021/acs.chemrev.8b00625Last update: Sep 2025